Support Vector Machine Kernel Similarity Space for Categorization
December 08, 2019
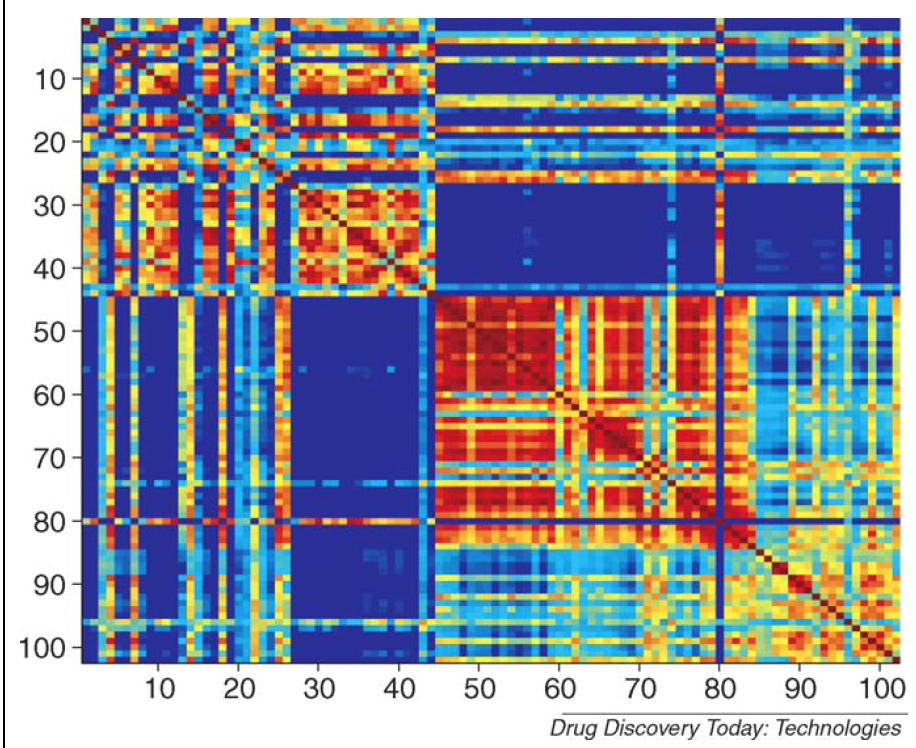
Illustration of SVM Similarity Matrix
Instead of working with the original sample representation in the original 6500 dimensional peptide space, SVM and GP classification methods operate directly on what is called the KERNEL MATRIX (see Glossary). This matrix has elements which correspond to the similarities between data points in the original space; in other words, each element of the matrix gives a measure of how similar the measured peptide profiles are between a pair of samples (red indicates highly similar and blue indicates dissimilarity). So the above figure shows how similar the peptide profiles are for every pair of samples available, where each axis corresponds to the total available samples (100 in total). The first thing to note is that we have now drastically reduced the dimensionality of the representation of each sample, from a point in a 6500 dimensional peptide profile space to a point in a 100 dimensional ‘peptide profile similarity space’. The second thing to note is that the matrix identifies that there are two groups of samples which all share highly similar peptide profiles (samples 1–45 and samples 46–86) with some evidence of a smaller group (samples 86–100). This data representation is now employed in SVM and GP classifiers rather than the original multidimensional space.
SVMs often are reported as achieving superior classification performance compared to other methods when compared across most applications and tasks. What is of importance is that they are fairly insensitive to the ‘Curse of Dimensionality’, primarily because of the use of the ‘kernel’ matrix [26]. This matrix is derived from the similarities between samples and thus operate on dimensions which are equal to the number of data points available. (Figure 3 gives an example derived from 100 samples, each of dimension 6500.) Therefore, SVMs are computationally reasonably efficient to cope with large-scale classification in both sample and variables. In clinical bioinformatics, they have the potential to provide powerful experimental disease diagnostic models based on gene or protein expression data with thousands of features and a small number, as little as a few dozen of samples [27,28].
Source: Girolami, Mark, Harald Mischak, and Ronald Krebs. "Analysis of complex, multidimensional datasets." Drug Discovery Today, Technologies 3.1 (2006), 13-19.
Source URL: https://www.sciencedirect.com/science/article/abs/pii/S1740674906000102?via%3Dihub
ID: support-vector-machine-kernel-similarity-space-for-categorization
Categories:
Tags: Boerhaave | COGWEB | Chinese | LSTM | Leiden | PGM | RNN | Ruysch | actors | aesthesis | agency | algorithm | analysis | anatomical | anatomy | androgynous | architecture | archive | artificialia | axis | black-box | body | botanical | brain | categories | categorization | channel | character recognition | chinese | classification | clustering | cnn | codes | cognition | collecting | collection | collections | colonialism | commodification | concept | conceptual-clustering | convolutional neural network | cost | counting | cut | cuts | cutting | datasets | demonstration | diagram | dimensionality | disgust | dissection | distance | domestication | elegance | epistemology | error | euclidean | evaluation | eye | figures | finger | forecasting | forensics | frame | freakish | geometry | gesture | gestures | gradient descent | graph | graphs | grouping | hacking | hand | hand writing | hands | hands-on | handwriting | hardware | history | human | human body | imagination | imperfect | inscription | instruments | joint | kmeans | knowledge | labeling | landmark | learning | location | machine learning | machines | materiality | meaning | measurement | memory | mnist | model | models | monsters | muscles | mystical | mythological | naturalia | nerves | nervous system | network | networks | neural networks | neural-anatomy | neuron | nonlinearity | observation | offline | online | ontologies | ontology | ontology-building | optimization | orientation | orthogonality | parallel | pca | perception | perceptron | perfection | performance | planes | poetic | position | prediction | preparation | preparations | projection | proportion | proportions | psychology | python | races | representation | representations | rhetoric | rnn | segments | selection | sensory experience | sensory perception | similarity | skeleton | skin | skull | skulls | space | sparseness | spectacle | spectators | speech | standard | statistic-ontology | statistical | statistical-ontology | svm | symbols | tacit | taxonomy | theatre | time-series | timeseries | tools | topological | training | treatise | trial | truth | type | typography | unsupervised | vision | visualization | wellcome | word2vec | writing | zodiac |